 |
POPSICLE- a software suite to determine population structure and to establish genotype-phenotype associations using Next-generation sequencing data |  |
MAIN INDEX ANALYTICAL PIPELINE CONTACT SYSTEM REQUIREMENTS POPSICLE Package Download Manual pages in HTML format Used Case |
FindAlleles Prerequisites Align the reads to the genome and sort them by genomic position using tools such as samtools. See here for details. What does it do? Finds the nucleotide composition at positions where there are reads aligned to the genome How to run it? java -jar LPDtools.jar findAlleles -i directoryBAMfiles -n 1 -o outputDirectoryAlleleFiles Here, -i is the directory where sorted aligned Binary Alignment map files are placed. -o is the output directory containing the allele files. -n is the bp window. -n 1 finds allele composition at each base Output The output directory contains the allele files that look as follows: 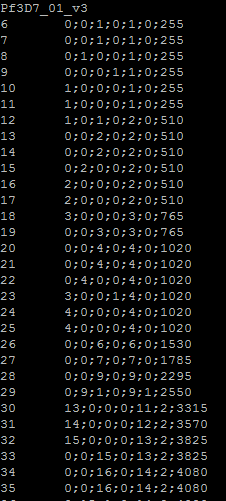 The chromosome name is listed first followed by other information corresponding to that chromosome. For example:
|
| CITATION: Jahangheer S. Shaik, Asis Khan and Michael E. Grigg, "POPSICLE: A Software Suite to Study Population Structure and Ancestral Determinants of Phenotypes using Whole genome Sequencing Data", submitted to PLoS special edition |