MAIN
INDEX
ANALYTICAL PIPELINE
CONTACT
SYSTEM REQUIREMENTS
POPSICLE Package
Download
Manual pages in HTML format
Used Case
|
Prerequisites
Align the reads to the genome and sort them by genomic position using
tools such as samtools. See here for details.
What does it do?
This utility finds the somies of the samples based on the cluster
assignments.
How to run it?
Run the utility as follows:
| java -jar LPDtools.jar FindSomies -i
directoryBAMfiles -o outputSomies.txt -m 1 -k chrSizes.txt |
Here,
-i is the directory where sorted aligned bam files are placed.
-m is the ploidy of the organism. For haploid organism, set m
to 1, for diploid, set m to 2
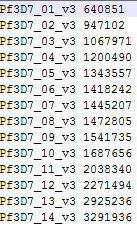
-k
is the file with chromosome sizes. Column 1 contains chromosome names
and column 2 contains the size of the chromosome. See below for an
example.
The
reads aligned to each chromosome are dependent on the somy of the
chomosome and the size of the chromosome. Higher the somy, more reads
are aligned to the chromosome. Bigger the chromosome, more reads are
aligned to the chromosome. The reads aligned to each chromosome are
normalized by the chromsosome sizes. The average value of these
normalized values correspond to the ploidy of the organism.
Accordingly, the normalized values are scaled and the somies are
reported.
Output
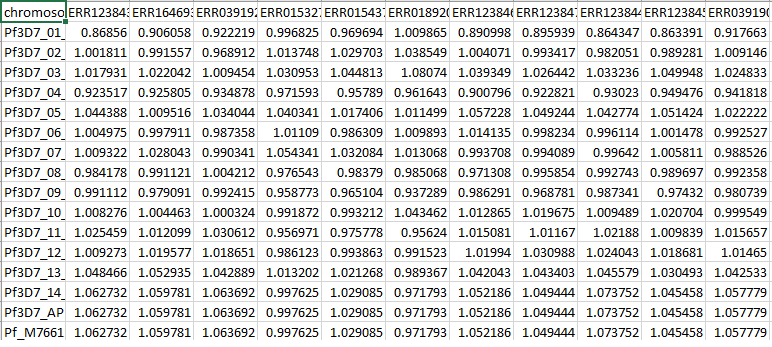
Here, each column is a sample and each row is a chromosome. Each block is therefore, somy of a chromosome in a sample. |
|
|