 |
POPSICLE- a software suite to determine population structure and to establish genotype-phenotype associations using Next-generation sequencing data |  |
MAIN INDEX ANALYTICAL PIPELINE CONTACT SYSTEM REQUIREMENTS POPSICLE Package Download Manual pages in HTML format Used Case |
Find regions with strong genotype-phenotype association Prerequisites
It finds regions with strong association between block genotypes and phenotypes How to run it? java -jar LPDtools.jar FindGenotypePhenotypeBootstrap -i localAncestryFile -j virulenceInformation -o outputFile -n bootstrapIterations Here, -i is the local ancestry inforamation found using the utility POPSICLEIntermediate of POPSICLE. -j is a file containing two columns, the first column contains the sample names and the second column contains phenotypes associated with the samples. Only categorical variables are accepted for phenotypes. -n is the number of bootstrapped datasets to be generated (-n 50 is sufficient for most purposes). Output 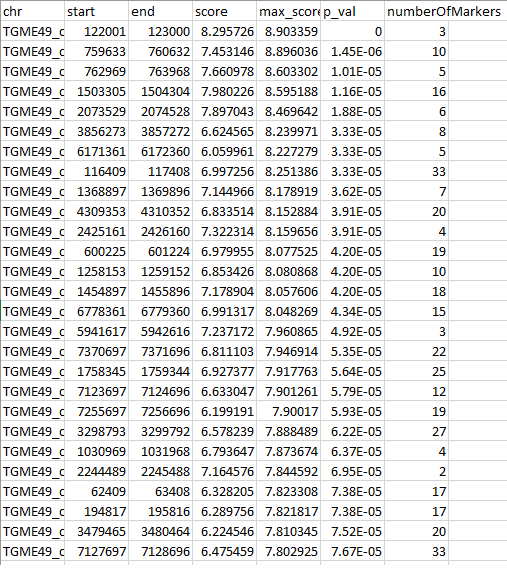 First column is chromosome name, columns 2 and 3 are start and end of the block respectively. column 4 is the score associated with original dataset. column 5 is the maximum scored recorded out of all bootstrapped datasets, column 5 is the p-value associated with a block and the last column is the number of markers in that block. |
| CITATION: Jahangheer S. Shaik, Asis Khan and Michael E. Grigg, "POPSICLE: A Software Suite to Study Population Structure and Ancestral Determinants of Phenotypes using Whole genome Sequencing Data", submitted to PLoS special edition |