 |
POPSICLE- a software suite to determine population structure and to establish genotype-phenotype associations using Next-generation sequencing data |  |
MAIN INDEX ANALYTICAL PIPELINE CONTACT SYSTEM REQUIREMENTS POPSICLE Package Download Manual pages in HTML format Used Case |
Find local Ancestries Prerequisites
It finds shared ancestries between different strains and species How to run it? java -jar LPDtools.jar POPSICLEIntermediate -i inputPOPSICLEbaselineFile -j clustersFile -o outputAncestryFile -n blockSize -k baselineARFFFile -l temporaryDirectory Here, -i is the baseline files generated using FindDivergenceFromBaseline utility of POPSICLE. -j is the clusters file generated PerformKmeansClustering using utility of POPSICLE, -n is the desired block size (eg. 1000 suggests block size of 1kb), -o is the output file and -l is a directory where temporary files are written. This directory with temporary files can be deleted later Output 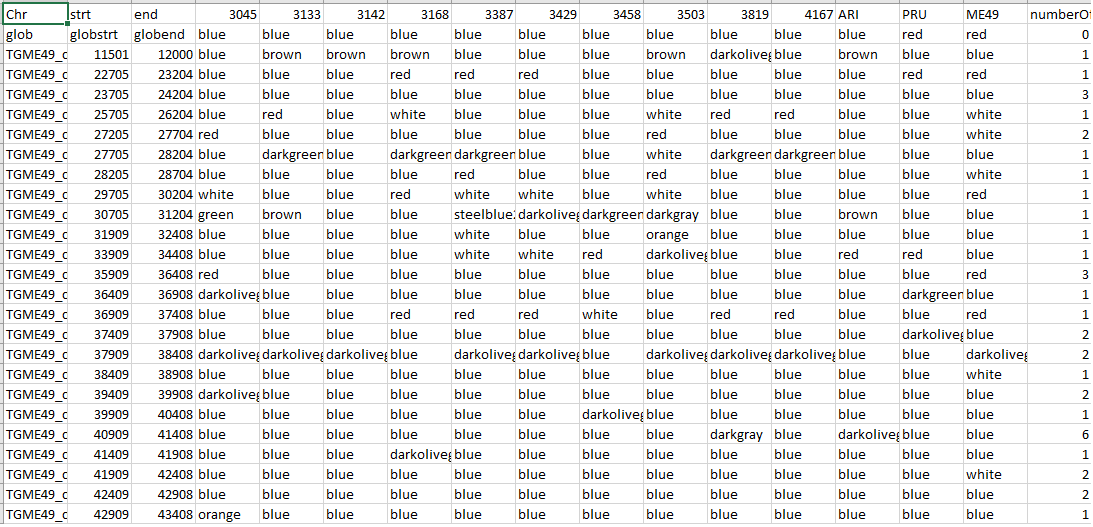 First column is chromosome name, columns 2 and 3 are start and end of the block columns 4 and greater are the local ancestry assignments of each sample. The last column is the number of markers in each block. The first row is header information, the second row is the clade assignment of each sample. |
| CITATION: Jahangheer S. Shaik, Asis Khan and Michael E. Grigg, "POPSICLE: A Software Suite to Study Population Structure and Ancestral Determinants of Phenotypes using Whole genome Sequencing Data", submitted to PLoS special edition |