 |
POPSICLE- a software suite to determine population structure and to establish genotype-phenotype associations using Next-generation sequencing data |  |
MAIN INDEX ANALYTICAL PIPELINE CONTACT SYSTEM REQUIREMENTS POPSICLE Package Download Manual pages in HTML format Used Case |
PerformKmeansClustering Prerequisites
It clusters the samples into specified range of groups using k-means implementation in WEKA. It also assigns score to each of those groups based on intra-cluster similarity and inter-cluster dissimilarity using DUNN index. It proposes optimum number of clusters that are relevant to your data. How to run it? java -jar LPDtools.jar PerformKmeansClustering -i sampledARFFfile -o outputClustersFile -n minClusters -m maxClusters Here, -i is the baseline files generated using FindDivergenceFromBaseline, which is converted into ARFF format using Convert2ARFFformat utility of POPSICLE Output 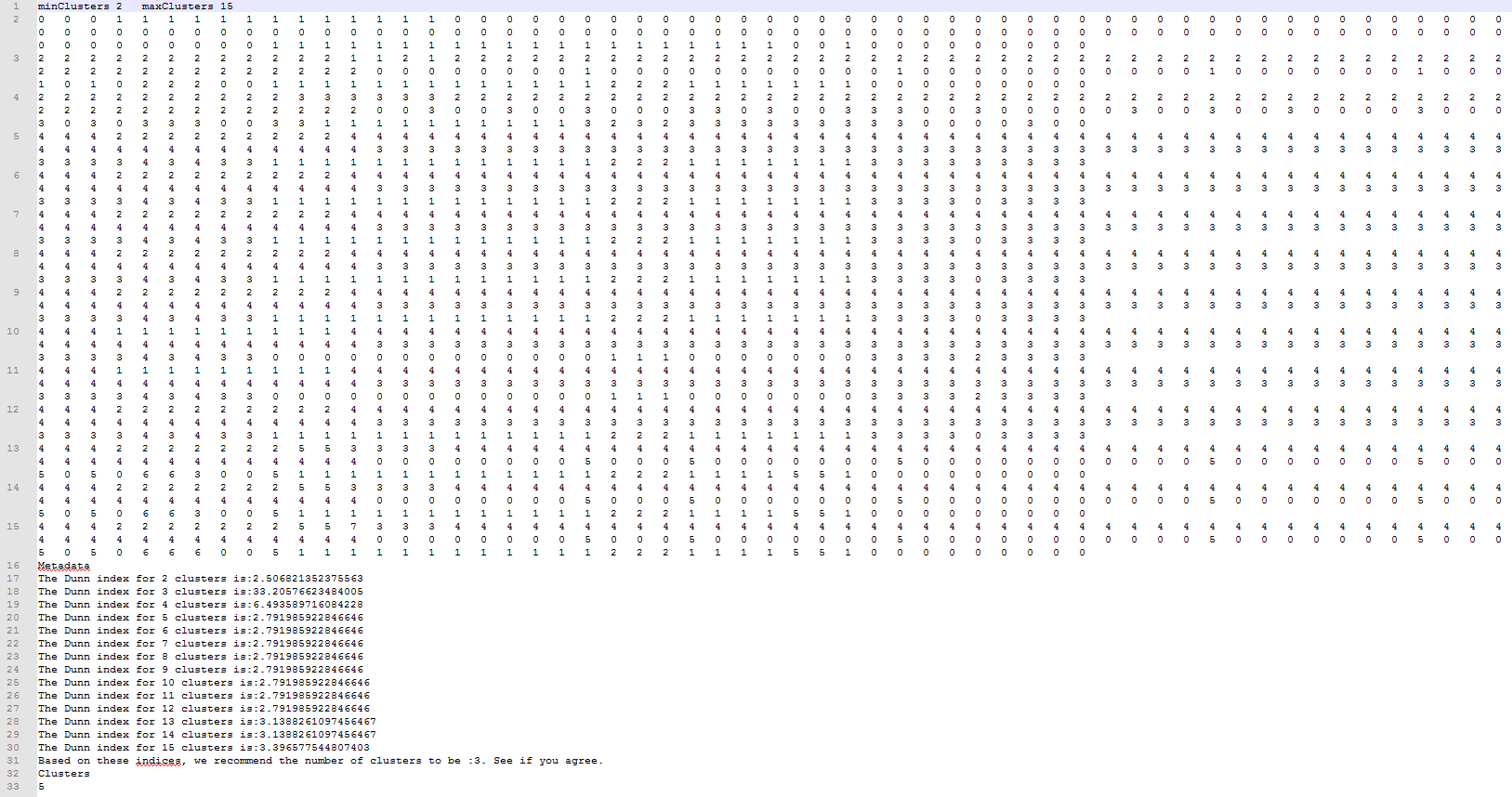 Here, the user specified minimum clusters to be 2 and maximum clusters to be 15 as indicated in the first line. The line 2 shows cluster assignments for each sample assigned to 2 clusters, line 3 shows assignments to 3 clusters and so on. Line 17 shows DUNN index score associated with 2 clusters, line 3 shows score for 3 clusters and so on. The software recommends the optimum number of clusters be 3. If the user disagrees with this evaluation, he can change the last line to reflect the number of clusters he thinks are relevant. In this example, the number of optimum clusters the user chose were 5 (line 33) |
| CITATION: Jahangheer S. Shaik, Asis Khan and Michael E. Grigg, "POPSICLE: A Software Suite to Study Population Structure and Ancestral Determinants of Phenotypes using Whole genome Sequencing Data", submitted to PLoS special edition |