 |
POPSICLE- a software suite to determine population structure and to establish genotype-phenotype associations using Next-generation sequencing data |  |
MAIN INDEX ANALYTICAL PIPELINE CONTACT SYSTEM REQUIREMENTS POPSICLE Package Download Manual pages in HTML format Used Case |
GenerateInputFromAlleleFiles Prerequisites
It generates a file that summarizes information from several allele files How to run it? java -jar LPDtools.jar GenerateInputFromAlleleFiles -i directoryAlleleFiles -j directoryOfSNPfiles -o outputPopsicleFile.txt -k "vcf" -l somiesFile Here, -i is the directory where allele files are present. -j is the directory where the Single nucleotide Polymorphism (SNP files) generated using a utility such as samtools are present. Alternately, one may use their own markers of choice. -k is the format of the snp files (VCF is the preferred format. Markers can be submitted in tab delimited format using "tab" tag). -l is the somies file Output 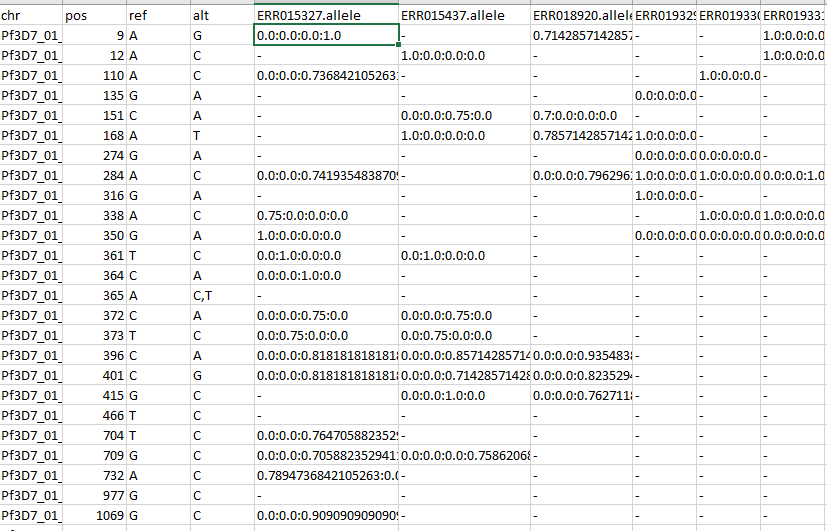 Based on the somy and the composition in the allele file, each nucleotide is given a weight. For example, the highlighted score corresponding to position 9 on chromosome pf3D7_01 gives 100% weight to the nucleotide "G". This signifies that this sample at position 9 contains alternate allele that is homozygous different from the reference. The missing data is represented by a "-" |
| CITATION: Jahangheer S. Shaik, Asis Khan and Michael E. Grigg, "POPSICLE: A Software Suite to Study Population Structure and Ancestral Determinants of Phenotypes using Whole genome Sequencing Data", submitted to PLoS special edition |